Metadata Retrieval
Query & retrieval of metadata (Analyses) from Song.
Using Swagger UI
Swagger UI is ideal for exploration and simple use cases. It provides detailed descriptions of all available endpoints, expected inputs, and error responses.
-
Locate your endpoint of interest: Use any
GETendpoint to retrieve data from Song. For example, under the Analysis dropdown, find theGETGetAnalysesForStudy endpoint.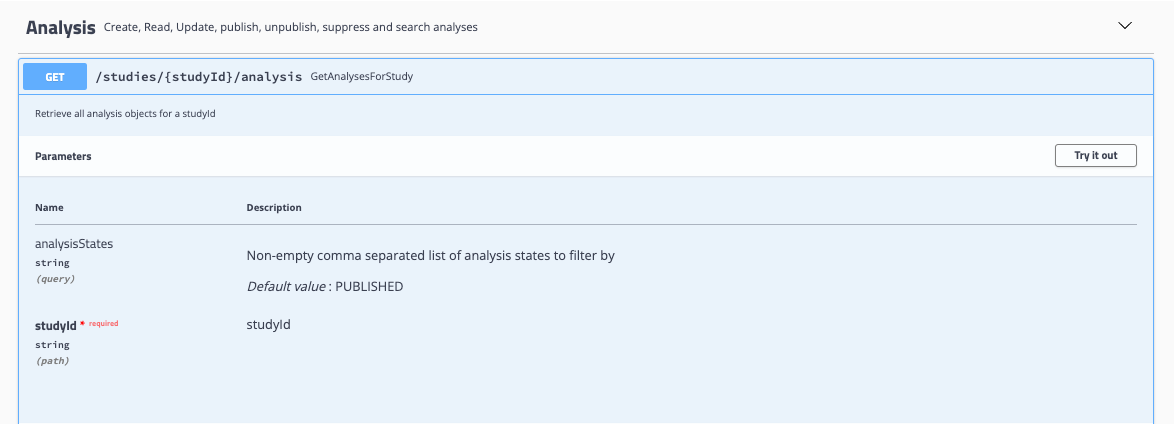
-
Select Try it out and input your desired values: For GetAnalysesForStudy, input a studyID and analysis state to return all associated analyses.
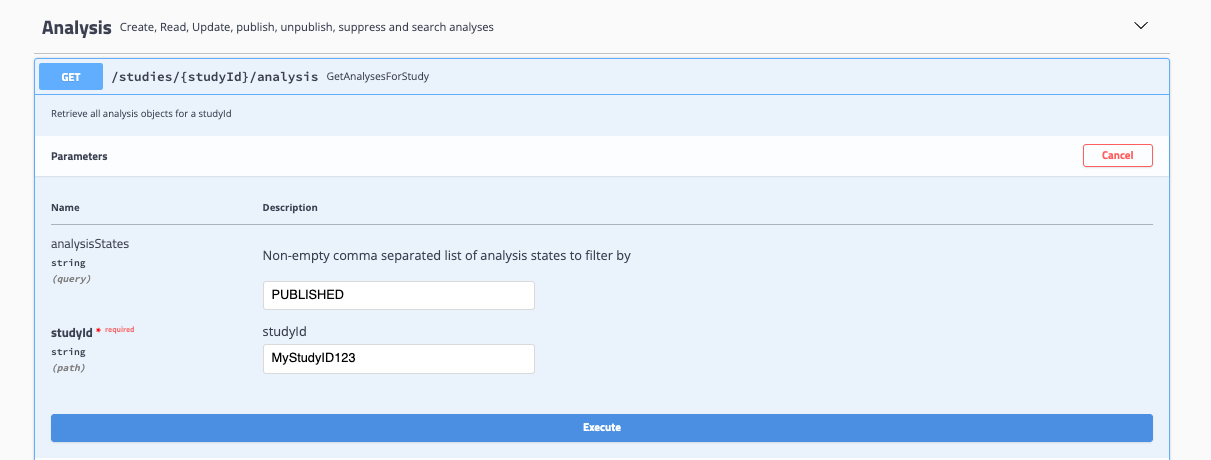
-
Select Execute: Expected responses, response codes, and descriptions are documented within Swagger-UI.
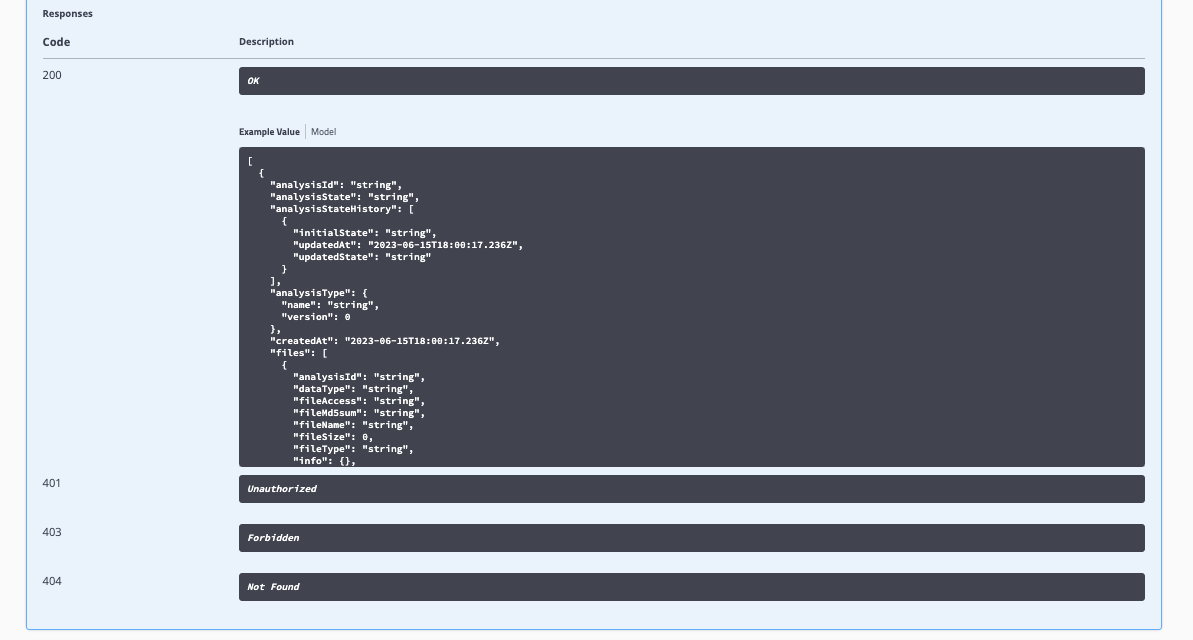
Programmatic Queries
Here are examples of how to retrieve analyses using the Song API programmatically.
Example 1: Sample ID Query
This Python script searches for associated analyses based on a specific sample ID:
import requests
url = "https://song.virusseq-dataportal.ca"
sample = "QC_546060"
study = "LSPQ-QC"
endpoint = f"{url}/studies/{study}/analysis/search/id?submitterSampleId={sample}"
response = requests.get(endpoint)
print(response.json()[0])
For technical support or specific use cases, please don't hesitate to reach out through our relevant community support channels.
Example 2: Bulk Analysis Query
This script demonstrates how to retrieve an aggregated list of published analyses and create a filtered list of analyses associated with a series of submitterIDs:
import requests
import pandas as pd
# Declare buckets to house analyses
aggregated_analyses = []
filtered_analyses = []
# Variables
study = "MCPL-MB"
status = "PUBLISHED"
url = "https://song.virusseq-dataportal.ca"
# Read CSV to identify specimens
tmp = pd.read_csv("/Users/esu/Desktop/GitHub/virus-seq/2023_05_31/DP_Update_consensus_seq_version.csv", sep=",")
# Clean up CSV
tmp.set_index('Specimen Collector Sample ID', inplace=True)
tmp['old'] = tmp["Consensus Sequence Software Version(old)"]
tmp['new'] = tmp["Consensus Sequence Software Version(new)"]
tmp.drop(["Consensus Sequence Software Version(old)", "Consensus Sequence Software Version(new)"], axis=1, inplace=True)
# First endpoint query
endpoint = f"{url}/studies/{study}/analysis/paginated?analysisStates={status}&limit=100&offset=0"
response = requests.get(endpoint)
# Check if query successful
if response.status_code != 200:
print(f"Error: {response.status_code}, {endpoint}")
exit(1)
# Get total analyses for pagination
total = response.json()['totalAnalyses']
# Append first set of analyses into bucket
aggregated_analyses.extend(response.json()['analyses'])
# Retrieve remaining pages of 100 queries and append to bucket
for offset in range(100, total, 100):
endpoint = f"{url}/studies/{study}/analysis/paginated?analysisStates={status}&limit=100&offset={offset}"
response = requests.get(endpoint)
if offset % 1000 == 0:
print(f"Retrieving {offset} out of {total} analyses")
if response.status_code != 200:
print(f"Error: {response.status_code}, {endpoint}")
exit(1)
aggregated_analyses.extend(response.json()['analyses'])
# Filter analyses according to sample specimen ID
print("Filtering analyses")
filtered_analyses = [analysis for analysis in aggregated_analyses
if analysis['samples'][0]['submitterSampleId'] in tmp.index.values]
print(f"Total analyses: {len(aggregated_analyses)}")
print(f"Filtered analyses: {len(filtered_analyses)}")
When making bulk calls, consider the time spent on API calls. For example, if you need to retrieve 800 out of 1000 analyses and only have submitter sample IDs, it's more efficient to make bulk calls of 100 analyses (totaling 10 API calls) and then filter for the desired submitter sample IDs, rather than making 800 individual API calls.